Mahdi Vasighi
Assistant Professor at Department of Computer Science and Information Technology
Institute for Advanced Studies in Basic Sciences (IASBS), Zanjan, Iran
Contact information
vasighi iasbs.ac.ir
vasighi gmail.com
+98 24 3315 3378
Institute for Advanced Studies in Basic Sciences (IASBS), No. 444, Prof. Yousef Sobouti Blvd., Zanjan 45137-66731, Iran
The International Conference on Contemporary Issues In Data Science

March 5-8, 2019

Institute for Advanced Studies in Basic Sciences (IASBS)
Research Interests
Another part of my researches is to design and develop more sophisticated multivariate data modeling and dimensionality reduction methods based on artificial neural networks. Most of the researches in this area are related to vector quantization based methods like the self-organizing map (SOM) which ease the exploratory data analysis in genomics, proteomics, and medical informatics.
Current Projects
Structural Classification of Proteins using Cellular Automata
Establishing structural and functional relationships between sequences in the presence of only the primary sequence information is a key task in biological sequence analysis. One of the main issues is how to interpret and represent protein sequences. There are many feature extraction and representation techniques usually convert the sequence to numerical values and vectors which could be used as input for many machine learning approaches.
Different approaches to extract features from sequences, feature selection and building a powerful classifier are the main subjects in structural classification of proteins.
Self Organizing Maps with Dynamic Structure
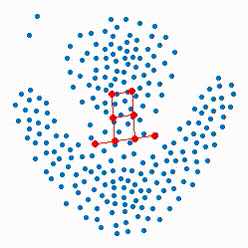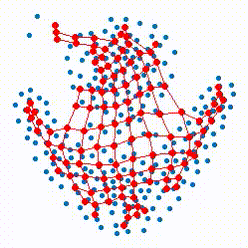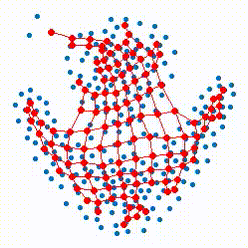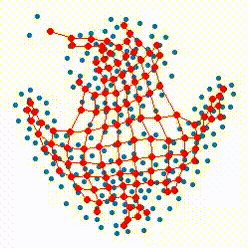
The growing self-organizing map (GSOM) possesses effective capability to generate feature maps and visualizing high-dimensional data without pre-determining their size by offering a flexible structure which enable the ability to learn the nonlinear manifolds in high dimensional feature space. Our recent researches focus on defining more reasonable strategies to control and direct the growth of the net in GSOMs. The proposed directed batch growing self organizing map (DBGSOM) uses a growing strategy based on the accumulative error around the candidate boundary neuron and helps to direct the network growth in proper directions.
Data Visualization and Dimensionality Reduction
In this project, we are working on nonlinear mapping for data visualization and dimensionaliy reduction which can resolve the deficiencies of classical SOMs. These deficencies like the static strcture and lack of topology preservation are the central subjects which considered to define more efficient mapping methods with an efficient topology preservation abilities.